Overview
brsmm() extends brs() to clustered data by
adding a Gaussian random intercept in the mean submodel while preserving
the interval-censored beta likelihood for scale-derived outcomes.
This vignette covers:
- full model mathematics;
- estimation by marginal maximum likelihood (Laplace approximation);
- practical use of all current
brsmmmethods; - inferential and validation workflows, including parameter recovery.
Mathematical model
Assume observations within groups , with random intercept .
Beta parameterization
For each
,
repar maps to beta shape parameters
via brs_repar().
Conditional contribution by censoring type
Each observation contributes:
where are interval endpoints on , is beta density, and is beta CDF.
Laplace approximation used by brsmm()
Define and , with curvature . Then
brsmm() maximizes the approximated
with stats::optim(), and computes group-level posterior
modes
and local standard deviations from the local curvature.
Simulating clustered scale data
The next helper simulates data from a known mixed model to illustrate fitting, inference, and recovery checks.
sim_brsmm_data <- function(seed = 3501L, g = 24L, ni = 12L,
beta = c(0.20, 0.65),
gamma = c(-0.15),
sigma_b = 0.55) {
set.seed(seed)
id <- factor(rep(seq_len(g), each = ni))
n <- length(id)
x1 <- rnorm(n)
b_true <- rnorm(g, mean = 0, sd = sigma_b)
eta_mu <- beta[1] + beta[2] * x1 + b_true[as.integer(id)]
eta_phi <- rep(gamma[1], n)
mu <- plogis(eta_mu)
phi <- plogis(eta_phi)
shp <- brs_repar(mu = mu, phi = phi, repar = 2)
y <- round(stats::rbeta(n, shp$shape1, shp$shape2) * 100)
list(
data = data.frame(y = y, x1 = x1, id = id),
truth = list(beta = beta, gamma = gamma, sigma_b = sigma_b, b = b_true)
)
}
sim <- sim_brsmm_data()
str(sim$data)
#> 'data.frame': 288 obs. of 3 variables:
#> $ y : num 18 29 12 43 56 55 3 21 4 4 ...
#> $ x1: num -0.3677 -2.0069 -0.0469 -0.2468 0.7634 ...
#> $ id: Factor w/ 24 levels "1","2","3","4",..: 1 1 1 1 1 1 1 1 1 1 ...Fitting brsmm()
fit_mm <- brsmm(
y ~ x1,
random = ~ 1 | id,
data = sim$data,
repar = 2,
int_method = "laplace",
method = "BFGS",
control = list(maxit = 1000)
)
fit_mm
#>
#> Call:
#> brsmm(formula = y ~ x1, random = ~1 | id, data = sim$data, repar = 2,
#> int_method = "laplace", method = "BFGS", control = list(maxit = 1000))
#>
#> Mixed beta interval model (Laplace)
#> Observations: 288 | Groups: 24
#> Log-likelihood: -1231.3848
#> Random SD: 0.4303
#> Convergence code: 0Core methods
Coefficients and random effects
coef(fit_mm, model = "random") is on the optimizer scale
();
use exp() to report
.
coef(fit_mm, model = "full")
#> (Intercept) x1 (phi)_(Intercept) (sd)_id
#> 0.01051336 0.62964563 -0.13626654 -0.84336505
coef(fit_mm, model = "mean")
#> (Intercept) x1
#> 0.01051336 0.62964563
coef(fit_mm, model = "precision")
#> (phi)_(Intercept)
#> -0.1362665
coef(fit_mm, model = "random")
#> (sd)_id
#> -0.843365
exp(coef(fit_mm, model = "random"))
#> (sd)_id
#> 0.4302602
head(ranef.brsmm(fit_mm))
#> 1 2 3 4 5 6
#> -0.37076507 0.45544785 -0.17418739 -0.45940282 0.02481745 0.42957923Variance-covariance, summary and likelihood criteria
vc <- vcov(fit_mm)
dim(vc)
#> [1] 4 4
sm <- summary(fit_mm)
sm
#>
#> Call:
#> brsmm(formula = y ~ x1, random = ~1 | id, data = sim$data, repar = 2,
#> int_method = "laplace", method = "BFGS", control = list(maxit = 1000))
#>
#> Mixed beta interval model (Laplace)
#> Observations: 288 | Groups: 24
#> logLik =-1231.3848 | AIC =2470.7696 | BIC =2485.4215
#>
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 0.01051 0.05998 0.175 0.8609
#> x1 0.62965 0.08650 7.280 3.35e-13 ***
#> (phi)_(Intercept) -0.13627 0.07704 -1.769 0.0769 .
#> (sd)_id -0.84337 0.26045 -3.238 0.0012 **
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
logLik(fit_mm)
#> 'log Lik.' -1231.385 (df=4)
AIC(fit_mm)
#> [1] 2470.77
BIC(fit_mm)
#> [1] 2485.421
nobs(fit_mm)
#> [1] 288Fitted values, prediction and residuals
head(fitted(fit_mm, type = "mu"))
#> [1] 0.3562286 0.1646701 0.4037733 0.3738710 0.5300708 0.3169583
head(fitted(fit_mm, type = "phi"))
#> [1] 0.465986 0.465986 0.465986 0.465986 0.465986 0.465986
head(predict(fit_mm, type = "response"))
#> 1 1 1 1 1 1
#> 0.3562286 0.1646701 0.4037733 0.3738710 0.5300708 0.3169583
head(predict(fit_mm, type = "link"))
#> 1 1 1 1 1 1
#> -0.5917708 -1.6238828 -0.3897673 -0.5156458 0.1204284 -0.7677855
head(predict(fit_mm, type = "precision"))
#> [1] 0.465986 0.465986 0.465986 0.465986 0.465986 0.465986
head(predict(fit_mm, type = "variance"))
#> [1] 0.10686447 0.06409816 0.11218166 0.10908334 0.11607513 0.10088398
head(residuals(fit_mm, type = "response"))
#> [1] -0.17622865 0.12532992 -0.28377333 0.05612904 0.02992923 0.23304166
head(residuals(fit_mm, type = "pearson"))
#> [1] -0.53908822 0.49503053 -0.84724816 0.16994500 0.08784679 0.73370665Diagnostic plotting methods
plot.brsmm() supports base and ggplot2 backends:
plot(fit_mm, which = 1:4, type = "pearson")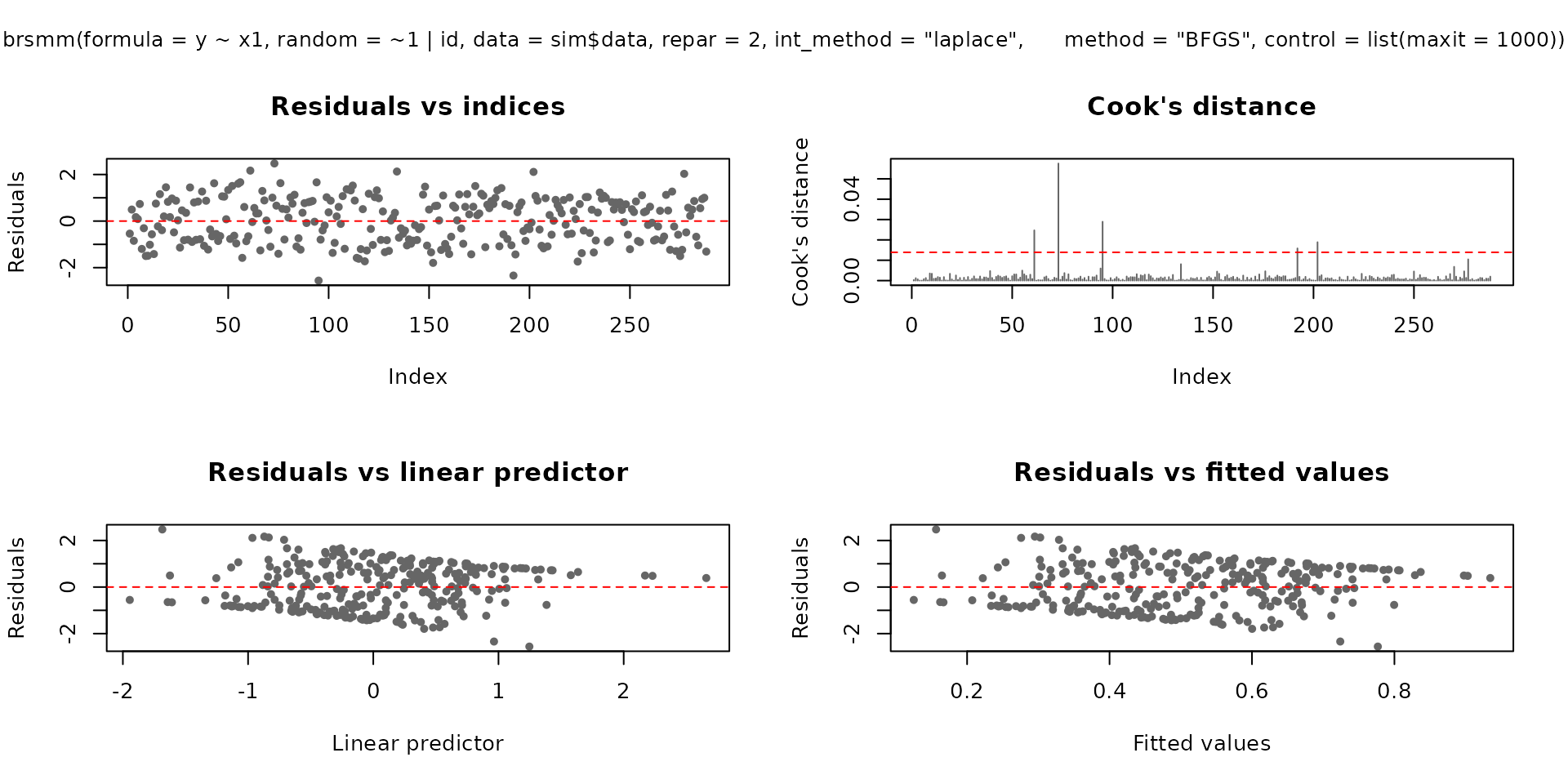
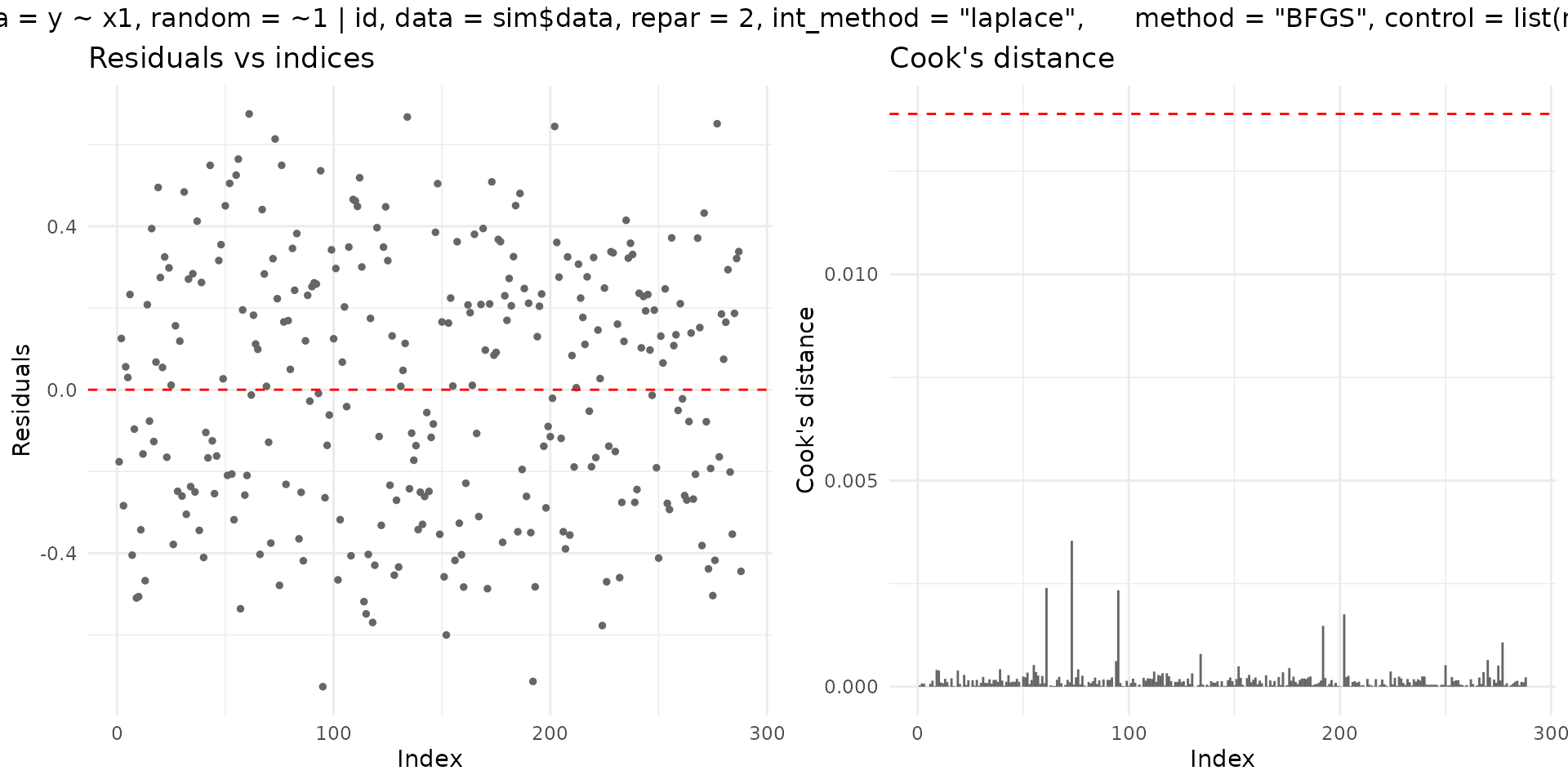
if (requireNamespace("ggplot2", quietly = TRUE)) {
plot(fit_mm, which = 1:2, gg = TRUE)
}
autoplot.brsmm() provides focused ggplot
diagnostics:
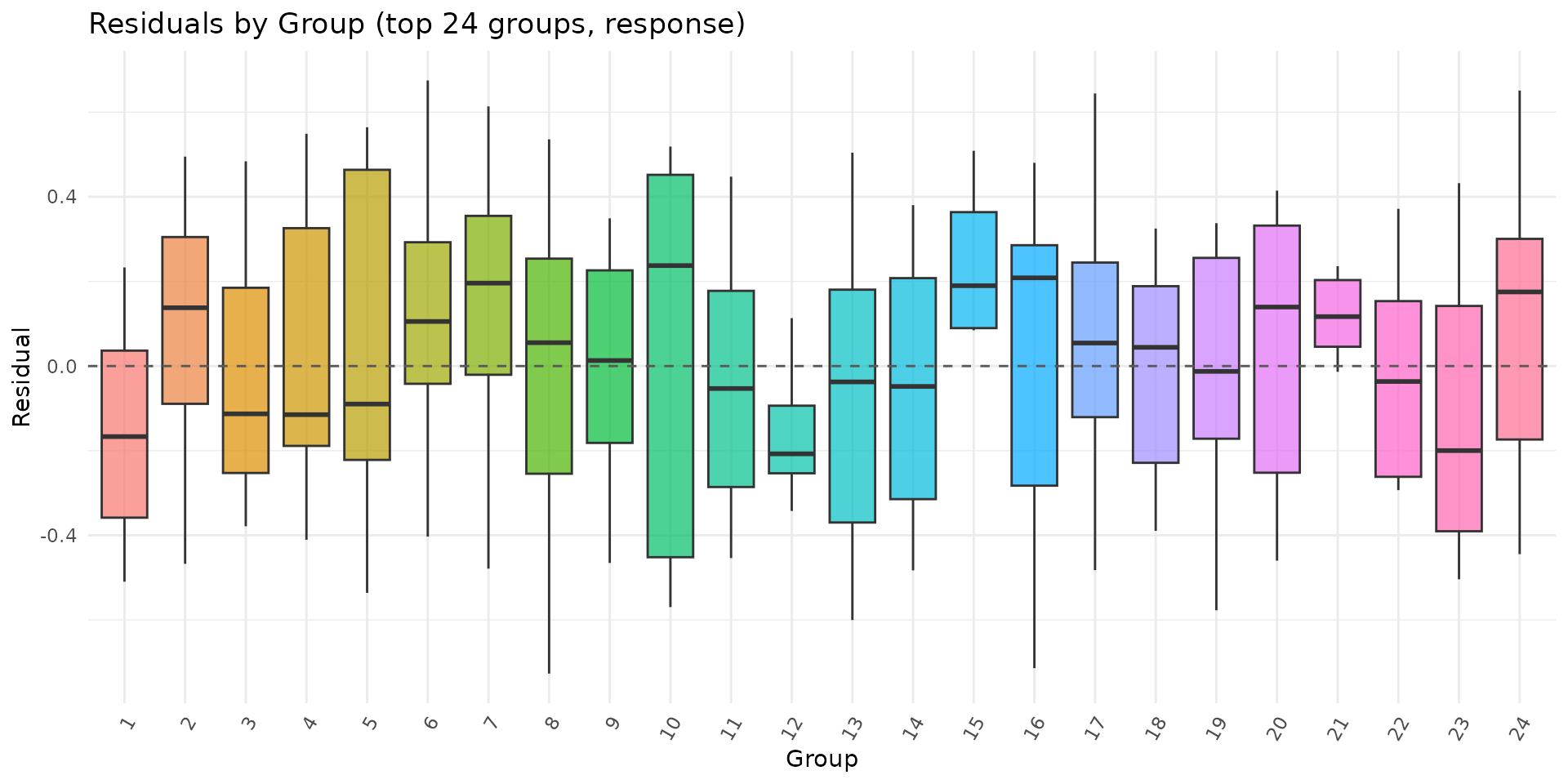
if (requireNamespace("ggplot2", quietly = TRUE)) {
autoplot.brsmm(fit_mm, type = "calibration")
autoplot.brsmm(fit_mm, type = "score_dist")
autoplot.brsmm(fit_mm, type = "ranef_qq")
autoplot.brsmm(fit_mm, type = "residuals_by_group")
}
Prediction with newdata
If newdata contains unseen groups,
predict.brsmm() uses random effect equal to zero for those
levels.
nd <- sim$data[1:8, c("x1", "id")]
predict(fit_mm, newdata = nd, type = "response")
#> [1] 0.3562286 0.1646701 0.4037733 0.3738710 0.5300708 0.3169583 0.4348323
#> [8] 0.3063863
nd_unseen <- nd
nd_unseen$id <- factor(rep("new_cluster", nrow(nd_unseen)))
predict(fit_mm, newdata = nd_unseen, type = "response")
#> [1] 0.4449724 0.2221609 0.4952496 0.4638431 0.6203876 0.4020284 0.5271241
#> [8] 0.3902401Statistical tests and validation workflow
Wald tests (from summary)
summary.brsmm() reports Wald
-tests
for each parameter:
summary(fit_mm)$coefficients
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 0.01051336 0.05997713 0.1752895 8.608522e-01
#> x1 0.62964563 0.08649567 7.2795044 3.350653e-13
#> (phi)_(Intercept) -0.13626654 0.07704204 -1.7687296 7.693901e-02
#> (sd)_id -0.84336505 0.26045257 -3.2380753 1.203390e-03Likelihood-ratio test for nested fixed effects
For nested models with the same random-effect structure, an LR test can be computed as: using a reference with parameter-count difference.
fit_null <- brsmm(
y ~ 1,
random = ~ 1 | id,
data = sim$data,
repar = 2,
int_method = "laplace",
method = "BFGS",
control = list(maxit = 1000)
)
ll0 <- logLik(fit_null)
ll1 <- logLik(fit_mm)
lr_stat <- 2 * (as.numeric(ll1) - as.numeric(ll0))
df_diff <- attr(ll1, "df") - attr(ll0, "df")
p_value <- stats::pchisq(lr_stat, df = df_diff, lower.tail = FALSE)
c(LR = lr_stat, df = df_diff, p_value = p_value)Residual diagnostics (quick checks)
r <- residuals(fit_mm, type = "pearson")
c(
mean = mean(r),
sd = stats::sd(r),
q025 = as.numeric(stats::quantile(r, 0.025)),
q975 = as.numeric(stats::quantile(r, 0.975))
)
#> mean sd q025 q975
#> 0.03803694 0.96104685 -1.56673066 1.62863050Parameter recovery experiment
A single-fit recovery table can be produced directly from the previous fit:
est <- c(
beta0 = coef(fit_mm, model = "mean")[1],
beta1 = coef(fit_mm, model = "mean")[2],
sigma_b = exp(coef(fit_mm, model = "random"))
)
true <- c(
beta0 = sim$truth$beta[1],
beta1 = sim$truth$beta[2],
sigma_b = sim$truth$sigma_b
)
recovery_table <- data.frame(
parameter = names(true),
true = as.numeric(true),
estimate = as.numeric(est[names(true)]),
bias = as.numeric(est[names(true)] - true)
)
recovery_table
#> parameter true estimate bias
#> 1 beta0 0.20 NA NA
#> 2 beta1 0.65 NA NA
#> 3 sigma_b 0.55 NA NAFor a Monte Carlo recovery study, repeat simulation and fitting across replicates:
mc_recovery <- function(R = 50L, seed = 7001L) {
set.seed(seed)
out <- vector("list", R)
for (r in seq_len(R)) {
sim_r <- sim_brsmm_data(seed = seed + r)
fit_r <- brsmm(
y ~ x1,
random = ~ 1 | id,
data = sim_r$data,
repar = 2,
int_method = "laplace",
method = "BFGS",
control = list(maxit = 1000)
)
out[[r]] <- c(
beta0 = coef(fit_r, model = "mean")[1],
beta1 = coef(fit_r, model = "mean")[2],
sigma_b = exp(coef(fit_r, model = "random"))
)
}
est <- do.call(rbind, out)
truth <- c(beta0 = 0.20, beta1 = 0.65, sigma_b = 0.55)
data.frame(
parameter = colnames(est),
truth = as.numeric(truth[colnames(est)]),
mean_est = colMeans(est),
bias = colMeans(est) - truth[colnames(est)],
rmse = sqrt(colMeans((sweep(est, 2, truth[colnames(est)], "-"))^2))
)
}
mc_recovery(R = 50)How this maps to automated package tests
The package test suite includes dedicated brsmm tests
for:
- fitting with Laplace integration;
- one- and two-part formulas;
- S3 methods (
coef,vcov,summary,predict,residuals,ranef); - parameter recovery under known DGP settings.
Run locally:
devtools::test(filter = "brsmm")References
Ferrari, S. L. P. and Cribari-Neto, F. (2004). Beta regression for modelling rates and proportions. Journal of Applied Statistics, 31(7), 799-815.
Lopes, J. E. (2024). Beta Regression for Interval-Censored Scale-Derived Outcomes. MSc Dissertation, PPGMNE/UFPR.
Pinheiro, J. C. and Bates, D. M. (2000). Mixed-Effects Models in S and S-PLUS. Springer.