Overview
The betaregscale package provides maximum-likelihood estimation of beta regression models for responses derived from bounded rating scales. Common examples include pain intensity scales (NRS-11, NRS-21, NRS-101), Likert-type scales, product quality ratings, and any instrument whose response can be mapped to the open interval .
The key idea is that a discrete score recorded on a bounded scale carries measurement uncertainty inherent to the instrument. For instance, a pain score of on a 0–10 NRS is not an exact value but rather represents a range: after rescaling to , the observation is treated as interval-censored in . The package uses the beta distribution to model such data, building a complete likelihood that supports mixed censoring types within the same dataset.
Installation
# Development version from GitHub:
# install.packages("remotes")
remotes::install_github("evandeilton/betaregscale")Censoring types
The complete likelihood (Lopes, 2024, Eq. 2.24) supports four
censoring types, automatically classified by
brs_check():
| Type | Likelihood contribution | |
|---|---|---|
| 0 | Exact (uncensored) | |
| 1 | Left-censored () | |
| 2 | Right-censored () | |
| 3 | Interval-censored |
where and are the beta density and CDF, are the interval endpoints, and are the beta shape parameters derived from and via the chosen reparameterization.
Interval construction
Scale observations are mapped to with midpoint uncertainty intervals:
where
is the number of scale categories (ncuts) and
is the half-width (lim, default 0.5).
# Illustrate brs_check with a 0-10 NRS scale
y_example <- c(0, 3, 5, 7, 10)
cr <- brs_check(y_example, ncuts = 10)
cr
#> left right yt y delta
#> [1,] 0.00001 0.05000 0.00001 0 1
#> [2,] 0.25000 0.35000 0.30000 3 3
#> [3,] 0.45000 0.55000 0.50000 5 3
#> [4,] 0.65000 0.75000 0.70000 7 3
#> [5,] 0.95000 0.99999 0.99999 10 2The delta column shows that
is left-censored
(),
is right-censored
(),
and all interior values are interval-censored
().
Data preparation with brs_prep()
In practice, analysts may want to supply their own censoring
indicators or interval endpoints rather than relying on the automatic
classification of brs_check(). The brs_prep()
function provides a flexible, validated bridge between raw analyst data
and brs().
It supports four input modes:
Mode 1: Score only (automatic)
# Equivalent to brs_check - delta inferred from y
d1 <- data.frame(y = c(0, 3, 5, 7, 10), x1 = rnorm(5))
brs_prep(d1, ncuts = 10)
#> brs_prep: n = 5 | exact = 0, left = 1, right = 1, interval = 3
#> left right yt y delta x1
#> 1 0.00001 0.05000 0.00001 0 1 -1.400043517
#> 2 0.25000 0.35000 0.30000 3 3 0.255317055
#> 3 0.45000 0.55000 0.50000 5 3 -2.437263611
#> 4 0.65000 0.75000 0.70000 7 3 -0.005571287
#> 5 0.95000 0.99999 0.99999 10 2 0.621552721Mode 2: Score + explicit censoring indicator
# Analyst specifies delta directly
d2 <- data.frame(
y = c(50, 0, 99, 50),
delta = c(0, 1, 2, 3),
x1 = rnorm(4)
)
brs_prep(d2, ncuts = 100)
#> Warning: Observation(s) 3: delta = 2 (right-censored) but y != 100.
#> brs_prep: n = 4 | exact = 1, left = 1, right = 1, interval = 1
#> left right yt y delta x1
#> 1 0.50000 0.50000 0.50000 50 0 1.1484116
#> 2 0.00001 0.00500 0.00001 0 1 -1.8218177
#> 3 0.98500 0.99999 0.99000 99 2 -0.2473253
#> 4 0.49500 0.50500 0.50000 50 3 -0.2441996Mode 3: Interval endpoints with NA patterns
When the analyst provides left and/or right
columns, censoring is inferred from the NA pattern:
d3 <- data.frame(
left = c(NA, 20, 30, NA),
right = c(5, NA, 45, NA),
y = c(NA, NA, NA, 50),
x1 = rnorm(4)
)
brs_prep(d3, ncuts = 100)
#> brs_prep: n = 4 | exact = 1, left = 1, right = 1, interval = 1
#> left right yt y delta x1
#> 1 1e-05 0.05000 0.025 NA 1 -0.2827054
#> 2 2e-01 0.99999 0.600 NA 2 -0.5536994
#> 3 3e-01 0.45000 0.375 NA 3 0.6289820
#> 4 5e-01 0.50000 0.500 50 0 2.0650249Mode 4: Analyst-supplied intervals
When the analyst provides y, left, and
right simultaneously, their endpoints are used directly
(rescaled by
):
Using brs_prep with brs()
Data processed by brs_prep() is automatically detected
by brs() - the internal brs_check() step is
skipped:
set.seed(42)
n <- 1000
dat <- data.frame(x1 = rnorm(n), x2 = rnorm(n))
sim <- brs_sim(
formula = ~ x1 + x2, data = dat,
beta = c(0.2, -0.5, 0.3), phi = 1 / 5,
link = "logit", link_phi = "logit",
repar = 2
)
prep <- brs_prep(sim, ncuts = 100)
#> brs_prep: n = 1000 | exact = 0, left = 71, right = 106, interval = 823
fit_prep <- brs(y ~ x1 + x2,
data = prep, repar = 2,
link = "logit", link_phi = "logit"
)
summary(fit_prep)
#>
#> Call:
#> brs(formula = y ~ x1 + x2, data = prep, link = "logit", link_phi = "logit",
#> repar = 2)
#>
#> Quantile residuals:
#> Min 1Q Median 3Q Max
#> -3.9434 -0.5715 0.2788 0.6560 1.3786
#>
#> Coefficients (mean model with logit link):
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) -5.11254 0.03821 -133.798 < 2e-16 ***
#> x1 -0.29458 0.02680 -10.990 < 2e-16 ***
#> x2 0.18828 0.02695 6.986 2.83e-12 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Phi coefficients (precision model with logit link):
#> Estimate Std. Error z value Pr(>|z|)
#> (phi) -4.88273 0.05962 -81.9 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#> ---
#> Log-likelihood: -4517.3759 on 4 Df
#> Pseudo R-squared: 0.1010
#> Number of iterations: 35 (BFGS)
#> Censoring: 823 interval | 71 left | 106 rightExample 1: Fixed dispersion model
Simulating data
We simulate 200 observations from a beta regression model with fixed dispersion, two covariates, and logit link for the mean.
set.seed(4255)
n <- 1000
dat <- data.frame(x1 = rnorm(n), x2 = rnorm(n))
sim_fixed <- brs_sim(
formula = ~ x1 + x2,
data = dat,
beta = c(0.3, -0.6, 0.4),
phi = 1 / 10,
link = "logit",
link_phi = "logit",
ncuts = 100,
repar = 2
)
head(sim_fixed)
#> left right yt y delta x1 x2
#> 1 0.16500 0.175 0.17000 17 3 1.95102377 0.2888356
#> 2 0.98500 0.995 0.99000 99 3 0.77253858 1.6102584
#> 3 0.00001 0.005 0.00001 0 1 0.72640816 1.0542434
#> 4 0.57500 0.585 0.58000 58 3 0.04873961 -0.4923176
#> 5 0.09500 0.105 0.10000 10 3 -0.54450108 -1.4098059
#> 6 0.06500 0.075 0.07000 7 3 0.36002855 -1.0115571Each observation is centered in its interval. For example, a score of 67 on a 0–100 scale yields with interval .
Fitting the model
fit_fixed <- brs(
y ~ x1 + x2,
data = sim_fixed,
link = "logit",
link_phi = "logit",
repar = 2
)
summary(fit_fixed)
#>
#> Call:
#> brs(formula = y ~ x1 + x2, data = sim_fixed, link = "logit",
#> link_phi = "logit", repar = 2)
#>
#> Quantile residuals:
#> Min 1Q Median 3Q Max
#> -3.4331 -0.6619 0.0070 0.7339 3.0637
#>
#> Coefficients (mean model with logit link):
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 0.30604 0.04312 7.097 1.28e-12 ***
#> x1 -0.60332 0.04530 -13.317 < 2e-16 ***
#> x2 0.44135 0.04385 10.066 < 2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Phi coefficients (precision model with logit link):
#> Estimate Std. Error z value Pr(>|z|)
#> (phi) 0.10991 0.04057 2.709 0.00674 **
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#> ---
#> Log-likelihood: -4035.2262 on 4 Df
#> Pseudo R-squared: 0.2393
#> Number of iterations: 39 (BFGS)
#> Censoring: 809 interval | 65 left | 126 rightThe summary output follows the betareg package style,
showing separate coefficient tables for the mean and precision
submodels, with Wald z-tests and
-values
based on the standard normal distribution.
Goodness of fit
brs_gof(fit_fixed)
#> logLik AIC BIC pseudo_r2
#> 1 -4035.226 8078.452 8098.083 0.239315Comparing link functions
The package supports several link functions for the mean submodel. We can compare them using information criteria:
links <- c("logit", "probit", "cauchit", "cloglog")
fits <- lapply(setNames(links, links), function(lnk) {
brs(y ~ x1 + x2, data = sim_fixed, link = lnk, repar = 2)
})
# Estimates
est_table <- do.call(rbind, lapply(names(fits), function(lnk) {
e <- brs_est(fits[[lnk]])
e$link <- lnk
e
}))
est_table
#> variable estimate se z_value p_value ci_lower
#> 1 (Intercept) 0.3060440 0.04312435 7.096780 1.276975e-12 0.22152185
#> 2 x1 -0.6033155 0.04530453 -13.316890 1.846396e-40 -0.69211077
#> 3 x2 0.4413494 0.04384507 10.066111 7.800150e-24 0.35541462
#> 4 (phi) 0.1099131 0.04056639 2.709462 6.739243e-03 0.03040443
#> 5 (Intercept) 0.1865515 0.02619220 7.122409 1.060568e-12 0.13521578
#> 6 x1 -0.3652474 0.02663952 -13.710732 8.757029e-43 -0.41745989
#> 7 x2 0.2668812 0.02611963 10.217650 1.652993e-24 0.21568767
#> 8 (phi) 0.1118451 0.04051860 2.760338 5.774155e-03 0.03243005
#> 9 (Intercept) 0.2765516 0.04086691 6.767127 1.313647e-11 0.19645391
#> 10 x1 -0.5595132 0.04925738 -11.358973 6.692662e-30 -0.65605589
#> 11 x2 0.4092917 0.04464584 9.167523 4.840110e-20 0.32178752
#> 12 (phi) 0.1098255 0.04056937 2.707105 6.787281e-03 0.03031104
#> 13 (Intercept) -0.1949445 0.02881644 -6.765044 1.332688e-11 -0.25142369
#> 14 x1 -0.3814461 0.02769930 -13.770966 3.810830e-43 -0.43573575
#> 15 x2 0.2779569 0.02703699 10.280617 8.617404e-25 0.22496541
#> 16 (phi) 0.1203423 0.04032397 2.984387 2.841474e-03 0.04130879
#> ci_upper link
#> 1 0.3905662 logit
#> 2 -0.5145203 logit
#> 3 0.5272841 logit
#> 4 0.1894218 logit
#> 5 0.2378873 probit
#> 6 -0.3130349 probit
#> 7 0.3180747 probit
#> 8 0.1912601 probit
#> 9 0.3566493 cauchit
#> 10 -0.4629705 cauchit
#> 11 0.4967960 cauchit
#> 12 0.1893401 cauchit
#> 13 -0.1384653 cloglog
#> 14 -0.3271565 cloglog
#> 15 0.3309485 cloglog
#> 16 0.1993758 cloglog
# Goodness of fit
gof_table <- do.call(rbind, lapply(fits, brs_gof))
gof_table
#> logLik AIC BIC pseudo_r2
#> logit -4035.226 8078.452 8098.083 0.2393150
#> probit -4035.685 8079.370 8099.001 0.2382638
#> cauchit -4035.500 8079.000 8098.631 0.1918217
#> cloglog -4037.563 8083.126 8102.757 0.1823572Residual diagnostics
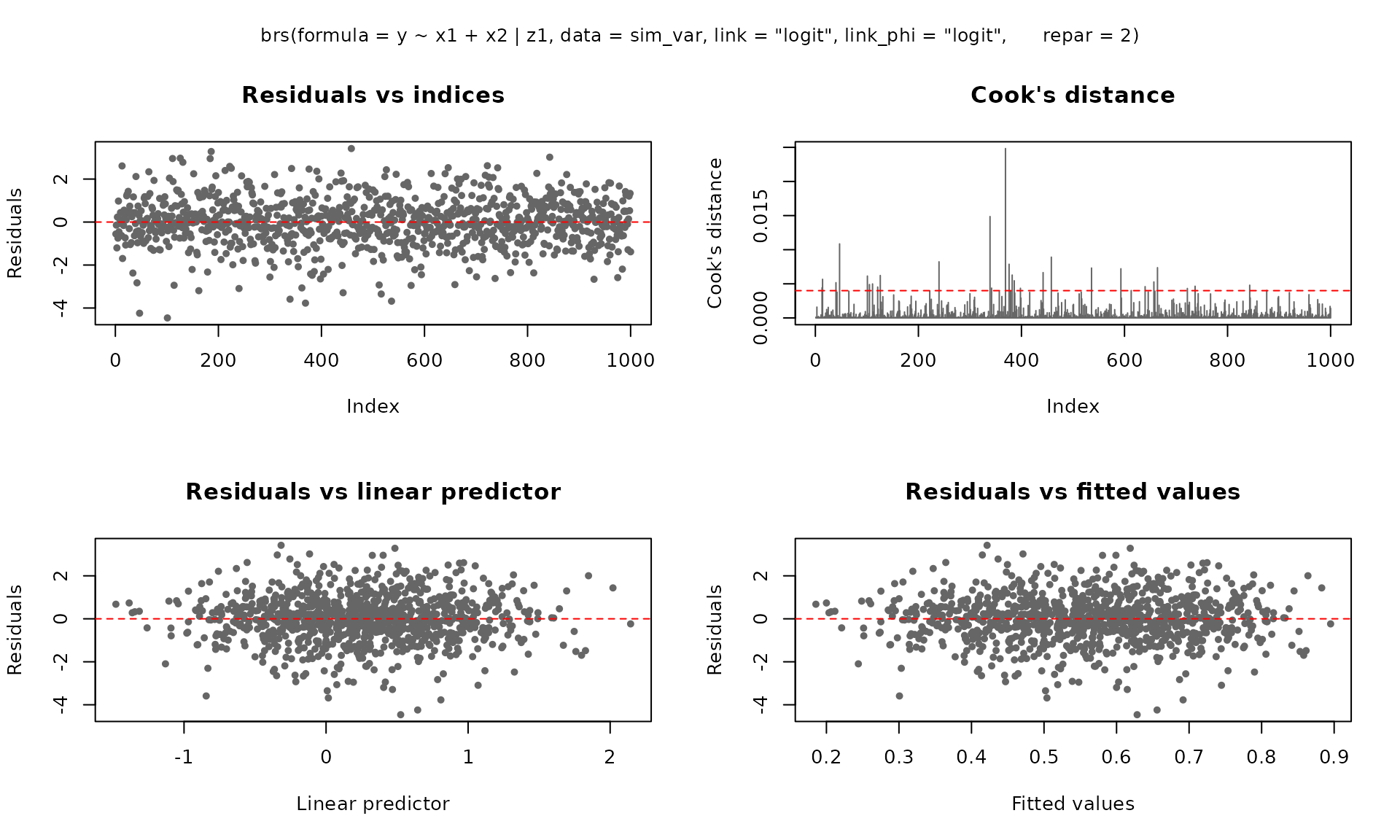
The plot() method provides six diagnostic panels. By
default, the first four are shown:
plot(fit_fixed)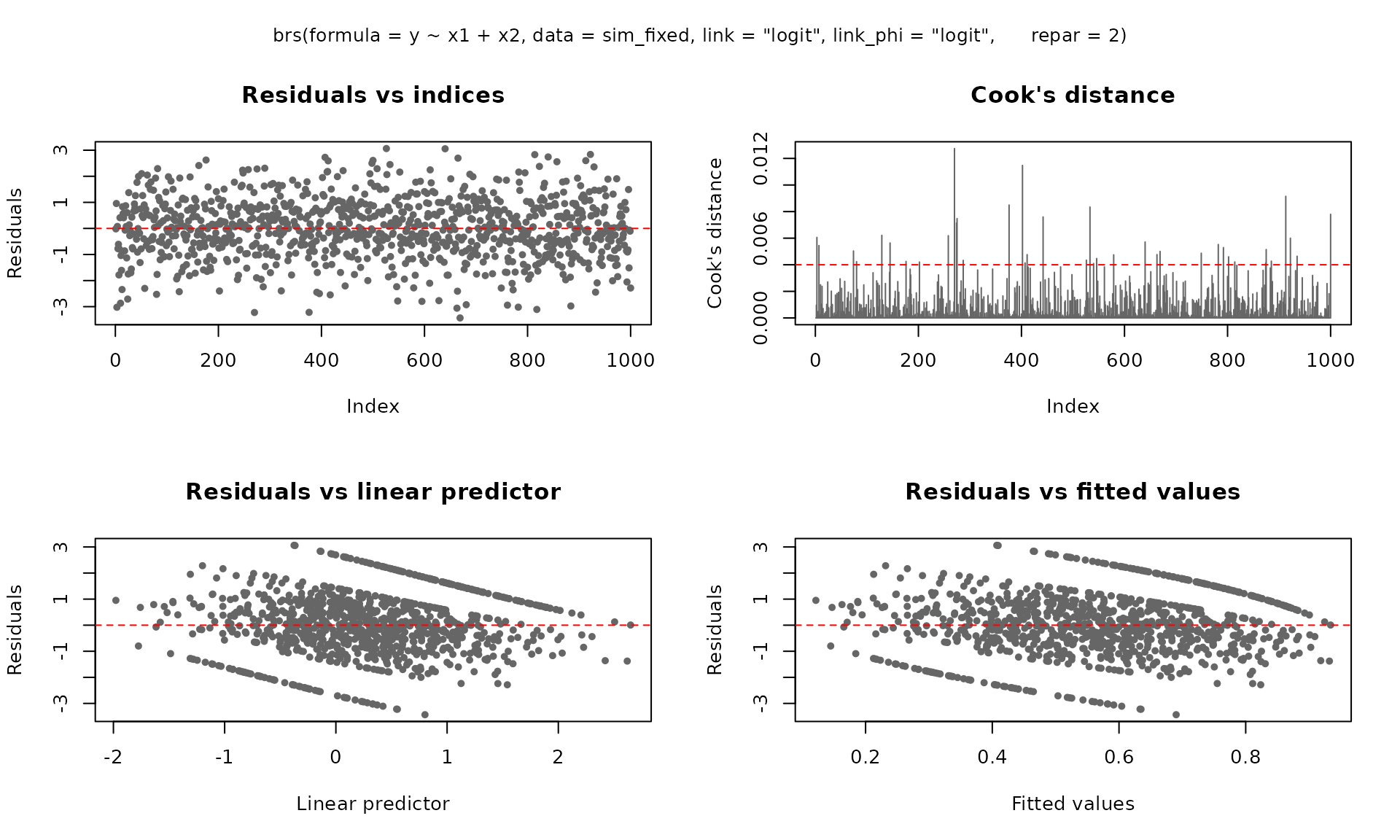
For ggplot2 output (requires the ggplot2 package):
plot(fit_fixed, gg = TRUE)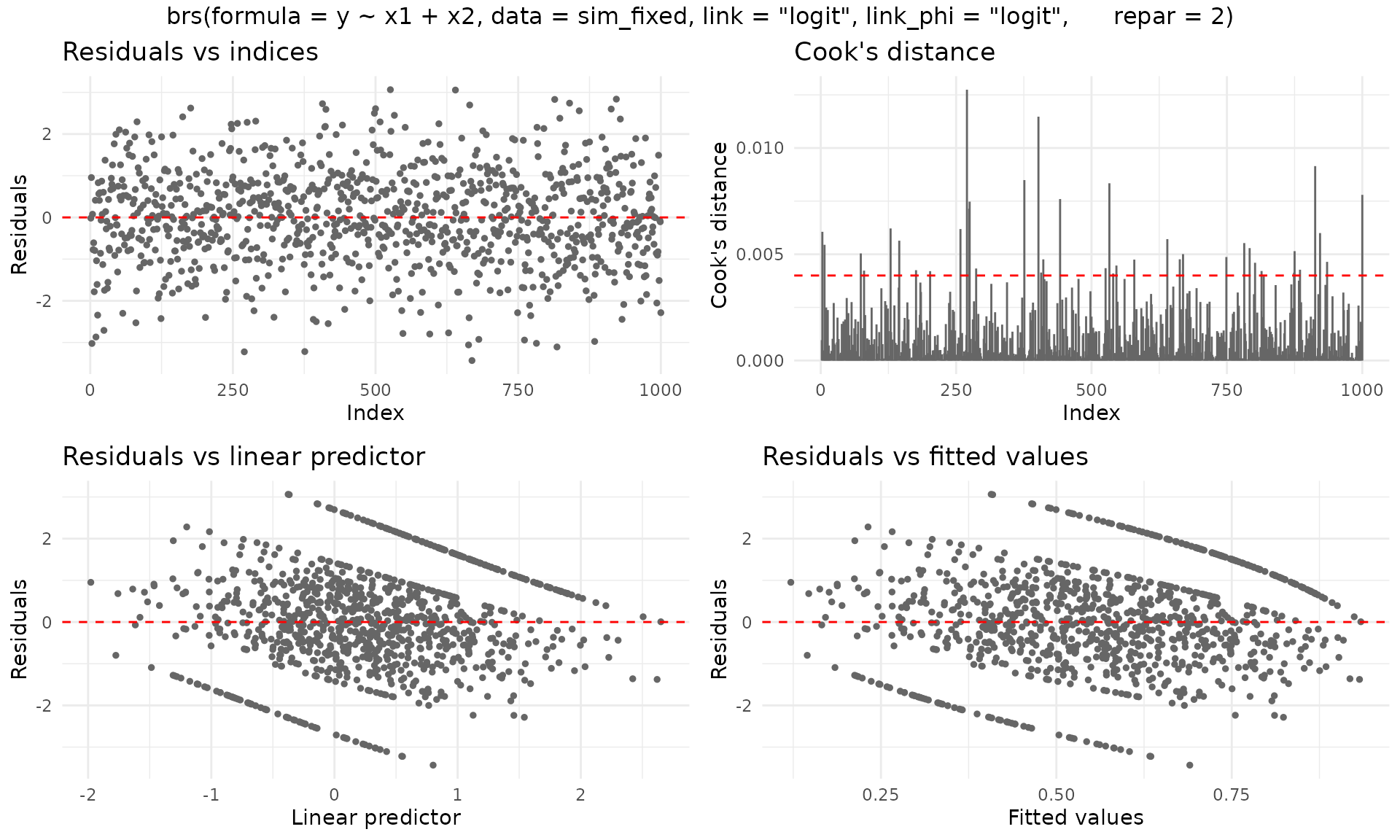
Predictions
# Fitted means
head(predict(fit_fixed, type = "response"))
#> [1] 0.3222258 0.6342855 0.5825087 0.5148343 0.5030832 0.4115366
# Conditional variance
head(predict(fit_fixed, type = "variance"))
#> [1] 0.1151933 0.1223514 0.1282719 0.1317466 0.1318576 0.1277349
# Quantile predictions
head(predict(fit_fixed, type = "quantile", at = c(0.25, 0.5, 0.75)))
#> q_0.25 q_0.5 q_0.75
#> [1,] 0.01699962 0.1779476 0.6024063
#> [2,] 0.31481409 0.7508330 0.9674995
#> [3,] 0.23110586 0.6578331 0.9397720
#> [4,] 0.14442537 0.5287586 0.8860292
#> [5,] 0.13183539 0.5059798 0.8744926
#> [6,] 0.05648038 0.3311318 0.7600605Censoring structure
The brs_cens() function provides a visual and tabular
overview of the censoring types in the fitted model:
brs_cens(fit_fixed)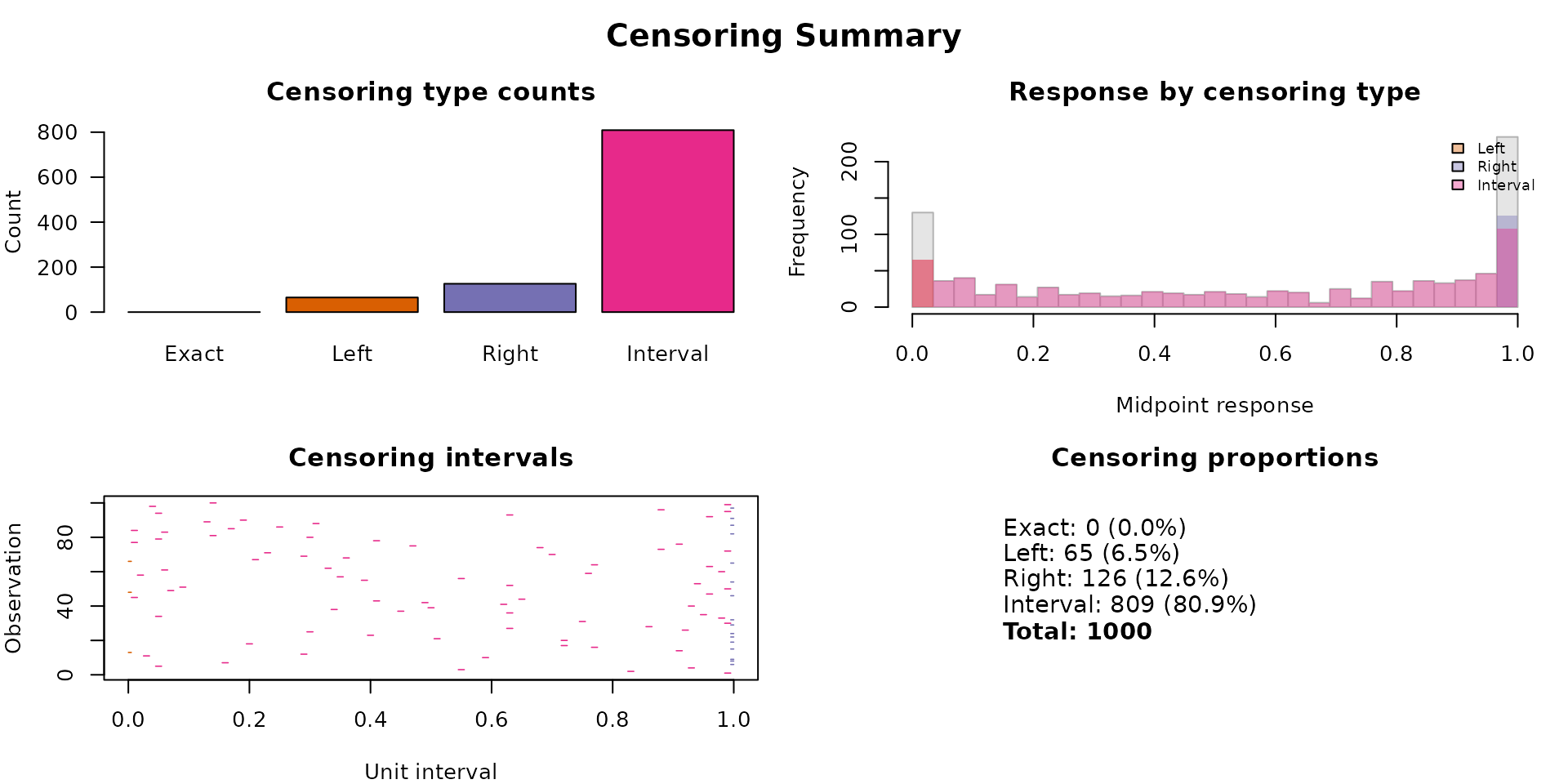
Example 2: Variable dispersion model
In many applications, the dispersion parameter
may depend on covariates. The package supports variable-dispersion
models using the Formula package notation:
y ~ x1 + x2 | z1 + z2, where the terms after |
define the linear predictor for
.
The same brs_sim() function is used for fixed and variable
dispersion; the second formula part activates the precision submodel in
simulation.
Simulating data
set.seed(2222)
n <- 1000
dat_z <- data.frame(
x1 = rnorm(n),
x2 = rnorm(n),
x3 = rbinom(n, size = 1, prob = 0.5),
z1 = rnorm(n),
z2 = rnorm(n)
)
sim_var <- brs_sim(
formula = ~ x1 + x2 + x3 | z1 + z2,
data = dat_z,
beta = c(0.2, -0.6, 0.2, 0.2),
zeta = c(0.2, -0.8, 0.6),
link = "logit",
link_phi = "logit",
ncuts = 100,
repar = 2
)
head(sim_var)
#> left right yt y delta x1 x2 x3 z1
#> 1 0.26500 0.275 0.27000 27 3 -0.3380621 -0.8235432 0 0.03062031
#> 2 0.17500 0.185 0.18000 18 3 0.9391643 -1.7562699 0 0.19384974
#> 3 0.00001 0.005 0.00001 0 1 1.7377190 -1.3148242 1 -0.42828641
#> 4 0.52500 0.535 0.53000 53 3 0.6963261 -0.8195514 0 0.36942900
#> 5 0.08500 0.095 0.09000 9 3 0.4622959 -0.1182560 1 0.05528553
#> 6 0.95500 0.965 0.96000 96 3 -0.3150868 -0.0648210 1 0.71694347
#> z2
#> 1 -1.1221809
#> 2 -0.2890842
#> 3 0.3479434
#> 4 0.2811058
#> 5 -1.2184445
#> 6 -0.5612588Fitting the model
fit_var <- brs(
y ~ x1 + x2 | z1,
data = sim_var,
link = "logit",
link_phi = "logit",
repar = 2
)
summary(fit_var)
#>
#> Call:
#> brs(formula = y ~ x1 + x2 | z1, data = sim_var, link = "logit",
#> link_phi = "logit", repar = 2)
#>
#> Quantile residuals:
#> Min 1Q Median 3Q Max
#> -4.4614 -0.6362 0.0056 0.6578 3.4202
#>
#> Coefficients (mean model with logit link):
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 0.27141 0.04245 6.394 1.62e-10 ***
#> x1 -0.54553 0.04375 -12.468 < 2e-16 ***
#> x2 0.17226 0.04141 4.160 3.18e-05 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Phi coefficients (precision model with logit link):
#> Estimate Std. Error z value Pr(>|z|)
#> (phi)_(Intercept) 0.24653 0.04233 5.823 5.77e-09 ***
#> (phi)_z1 -0.72230 0.04507 -16.028 < 2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#> ---
#> Log-likelihood: -3922.2430 on 5 Df
#> Pseudo R-squared: 0.1159
#> Number of iterations: 42 (BFGS)
#> Censoring: 744 interval | 105 left | 151 rightNotice the (phi)_ prefix in the precision coefficient
names, following the betareg convention.
Accessing coefficients by submodel
# Full parameter vector
coef(fit_var)
#> (Intercept) x1 x2 (phi)_(Intercept)
#> 0.2714053 -0.5455337 0.1722582 0.2465266
#> (phi)_z1
#> -0.7223034
# Mean submodel only
coef(fit_var, model = "mean")
#> (Intercept) x1 x2
#> 0.2714053 -0.5455337 0.1722582
# Precision submodel only
coef(fit_var, model = "precision")
#> (phi)_(Intercept) (phi)_z1
#> 0.2465266 -0.7223034
# Variance-covariance matrix for the mean submodel
vcov(fit_var, model = "mean")
#> (Intercept) x1 x2
#> (Intercept) 1.801851e-03 -7.620050e-05 4.312471e-05
#> x1 -7.620050e-05 1.914399e-03 2.137302e-05
#> x2 4.312471e-05 2.137302e-05 1.714705e-03Comparing link functions (variable dispersion)
links <- c("logit", "probit", "cauchit", "cloglog")
fits_var <- lapply(setNames(links, links), function(lnk) {
brs(y ~ x1 + x2 | z1, data = sim_var, link = lnk, repar = 2)
})
# Estimates
est_var <- do.call(rbind, lapply(names(fits_var), function(lnk) {
e <- brs_est(fits_var[[lnk]])
e$link <- lnk
e
}))
est_var
#> variable estimate se z_value p_value ci_lower
#> 1 (Intercept) 0.2714053 0.04244822 6.393797 1.618165e-10 0.18820830
#> 2 x1 -0.5455337 0.04375384 -12.468246 1.112475e-35 -0.63128964
#> 3 x2 0.1722582 0.04140899 4.159922 3.183568e-05 0.09109804
#> 4 (phi)_(Intercept) 0.2465266 0.04233361 5.823424 5.765393e-09 0.16355422
#> 5 (phi)_z1 -0.7223034 0.04506591 -16.027712 8.184127e-58 -0.81063101
#> 6 (Intercept) 0.1675402 0.02604987 6.431519 1.263347e-10 0.11648343
#> 7 x1 -0.3351860 0.02618407 -12.801144 1.615539e-37 -0.38650587
#> 8 x2 0.1056477 0.02529232 4.177066 2.952932e-05 0.05607566
#> 9 (phi)_(Intercept) 0.2467114 0.04232425 5.829080 5.573393e-09 0.16375741
#> 10 (phi)_z1 -0.7228855 0.04508355 -16.034353 7.354594e-58 -0.81124759
#> 11 (Intercept) 0.2313768 0.03790774 6.103682 1.036526e-09 0.15707898
#> 12 x1 -0.4748684 0.04383936 -10.832010 2.427663e-27 -0.56079197
#> 13 x2 0.1487280 0.03733542 3.983564 6.788945e-05 0.07555194
#> 14 (phi)_(Intercept) 0.2487158 0.04230585 5.878993 4.127710e-09 0.16579785
#> 15 (phi)_z1 -0.7182945 0.04497995 -15.969215 2.094062e-57 -0.80645363
#> 16 (Intercept) -0.2030447 0.02869977 -7.074785 1.496803e-12 -0.25929522
#> 17 x1 -0.3596966 0.02798194 -12.854597 8.104674e-38 -0.41454021
#> 18 x2 0.1150788 0.02705914 4.252863 2.110551e-05 0.06204385
#> 19 (phi)_(Intercept) 0.2490380 0.04225119 5.894224 3.764457e-09 0.16622719
#> 20 (phi)_z1 -0.7243353 0.04514341 -16.045206 6.175324e-58 -0.81281476
#> ci_upper link
#> 1 0.3546022 logit
#> 2 -0.4597777 logit
#> 3 0.2534183 logit
#> 4 0.3294989 logit
#> 5 -0.6339759 logit
#> 6 0.2185970 probit
#> 7 -0.2838662 probit
#> 8 0.1552197 probit
#> 9 0.3296654 probit
#> 10 -0.6345233 probit
#> 11 0.3056746 cauchit
#> 12 -0.3889448 cauchit
#> 13 0.2219041 cauchit
#> 14 0.3316338 cauchit
#> 15 -0.6301354 cauchit
#> 16 -0.1467942 cloglog
#> 17 -0.3048530 cloglog
#> 18 0.1681137 cloglog
#> 19 0.3318488 cloglog
#> 20 -0.6358559 cloglog
# Goodness of fit
gof_var <- do.call(rbind, lapply(fits_var, brs_gof))
gof_var
#> logLik AIC BIC pseudo_r2
#> logit -3922.243 7854.486 7879.025 0.11589802
#> probit -3922.227 7854.455 7878.994 0.12161749
#> cauchit -3923.161 7856.323 7880.862 0.09022080
#> cloglog -3922.296 7854.593 7879.132 0.08976588S3 methods reference
The following standard S3 methods are available for objects of class
"brs":
| Method | Description |
|---|---|
print() |
Compact display of call and coefficients |
summary() |
Detailed output with Wald tests and goodness-of-fit |
coef(model=) |
Extract coefficients (full, mean, or precision) |
vcov(model=) |
Variance-covariance matrix (full, mean, or precision) |
confint(model=) |
Wald confidence intervals |
logLik() |
Log-likelihood value |
AIC(), BIC()
|
Information criteria |
nobs() |
Number of observations |
formula() |
Model formula |
model.matrix(model=) |
Design matrix (mean or precision) |
fitted() |
Fitted mean values |
residuals(type=) |
Residuals: response, pearson, rqr, weighted, sweighted |
predict(type=) |
Predictions: response, link, precision, variance, quantile |
plot(gg=) |
Diagnostic plots (base R or ggplot2) |
Reparameterizations
The package supports three reparameterizations of the beta
distribution, controlled by the repar argument:
Direct (repar = 0): Shape parameters
and
are used directly. This is rarely used in practice.
Precision (repar = 1, Ferrari &
Cribari-Neto, 2004): The mean
and precision
yield
and
.
Higher
means less variability.
Mean–variance (repar = 2): The mean
and dispersion
yield
and
.
Here
acts as a coefficient of variation: smaller
means less variability.