Produces ggplot2 diagnostics tailored to mixed beta interval models.
Arguments
- object
A fitted
"brsmm"object.- type
Plot type:
"calibration","score_dist","ranef_qq", or"residuals_by_group".- bins
Number of bins used in calibration plots.
- scores
Optional integer vector of scores for
"score_dist". Defaults to all scores from0toncuts.- residual_type
Residual type passed to
residuals.brsmmfortype = "residuals_by_group".- max_groups
Maximum number of groups displayed in
"residuals_by_group".- ...
Currently ignored.
Examples
# \donttest{
if (requireNamespace("ggplot2", quietly = TRUE)) {
set.seed(123)
g <- 10
ni <- 8
id <- factor(rep(seq_len(g), each = ni))
n <- length(id)
x1 <- rnorm(n)
b <- rnorm(g, sd = 0.4)
mu <- plogis(0.1 + 0.5 * x1 + b[as.integer(id)])
phi <- plogis(-0.2)
shp <- brs_repar(mu = mu, phi = rep(phi, n), repar = 2)
y <- round(stats::rbeta(n, shp$shape1, shp$shape2) * 100)
d <- data.frame(y = y, x1 = x1, id = id)
fit_mm <- brsmm(y ~ x1, random = ~ 1 | id, data = d, repar = 2)
autoplot.brsmm(fit_mm, type = "calibration")
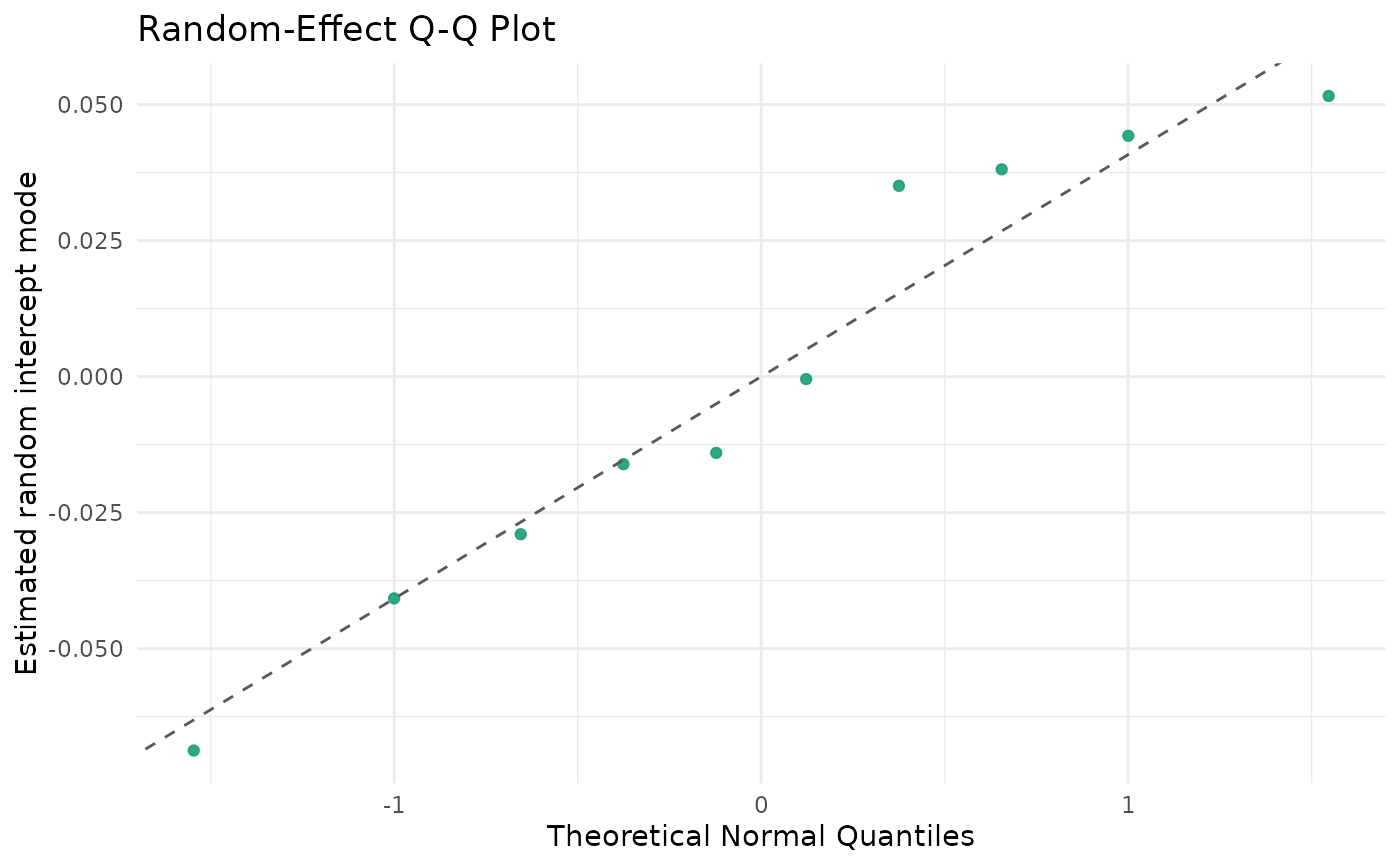
autoplot.brsmm(fit_mm, type = "ranef_qq")
}
 # }
# }